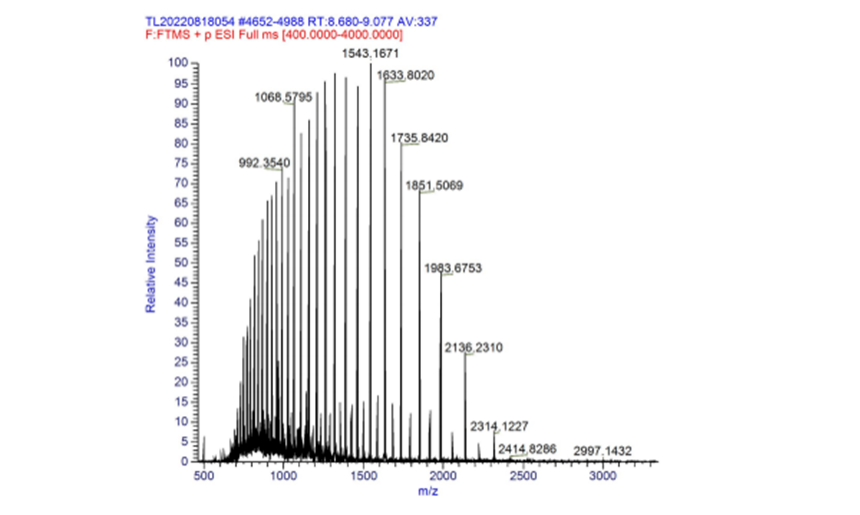
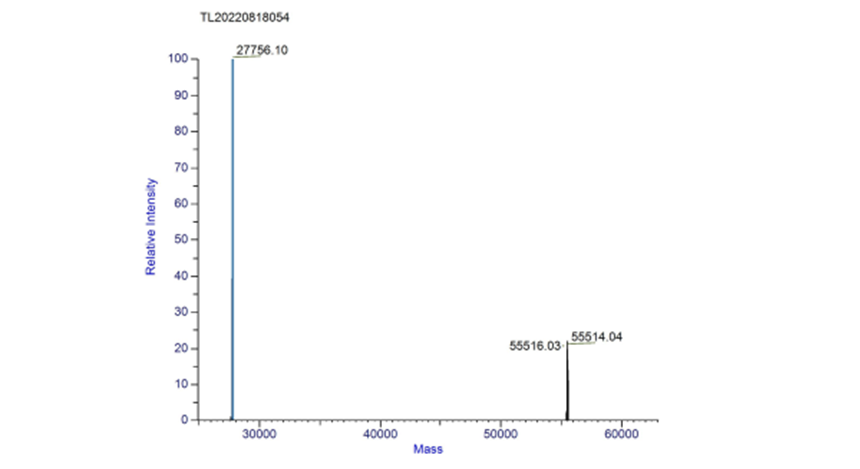
Part:BBa_K3468088
PETase A202C&E231C
The PETase is an enzyme, which can hydrolyze PET and this mutation protein is changed on the basis of the PETase. This protein is changed A202C/E231C which can be more stable in higher temperature compared with the wild type.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Attempts to stabilize folded proteins have naturally focused on the use of disulfide bonds. These covalent crosslinks have a reputation for being able to stabilize folded conformations by between 2 and 5 kcal/mol for each disulfide. Disulfide bonds provide stability to many extracellular and secreted proteins. Disulfide bonds are believed to decrease the conformational entropy and raise the free energy of the denatured state, thus providing an increase in stability to the folded protein conformation. While the overall effect of a disulfide bond may be complex, including an enthalpic component, considerable evidence supports the long-standing hypothesis that stability is gained through a reduction in unfolded conformational entropy. In our design two parallel folds (β7-β8) of PETase were introduced to stabilize this region and improve the thermal stability of the protein
We builded a model of the mutant (A202C-E231c), using Pymol for visual inspection (geometric criteria, see Figure 4 and Table 1), and finally using FoldX for evaluation. During visual inspection in Pymol, we found that this disulfide bond is far from the catalytic triad, far from the inside of the protein, and met the disulfide bond geometry criteria. DDG is -2.454kcal/mol evaluated by FoldX.
| None |